MSc AI student combines geometry and topology in ICML paper
29 June 2023

In this work, Floor combined two fundamentally different approaches to improving the expressivity of graph neural networks to leverage both benefits when doing e.g. molecular prediction tasks and modelling N-body systems. Floor was supervised in this project by Dr. Erik Bekkers and Rob Hesselink (AMLAB of the Informatics Institute).
The main ideas behind the paper
How do we learn the structure of graphs?
Graph neural networks (GNNs) have recently arisen as the most studied and adopted framework for learning data which can be described as graphs. Since graphs are a flexible way of representing many different types of data, GNNs are being developed in many scientific and industrial fields, e.g. learning on molecules and social networks. GNNs typically aim to learn functions that model complex interactions among the underlying topology and the provided features.
The learning procedure typically is done through ‘message passing’, i.e. updating the hidden representation of each node in the graph by aggregating the information of its neighbors. However, learning with this procedure is inherently limited, since graphs that look equivalent locally can have vastly different global topologies. Another way to view this is that local information aggregation is not powerful enough the distinguish all types of non-isomorphic graphs. Since the topology is an essential component in learning on graphs, this limitation constrains the performance of GNNs significantly.
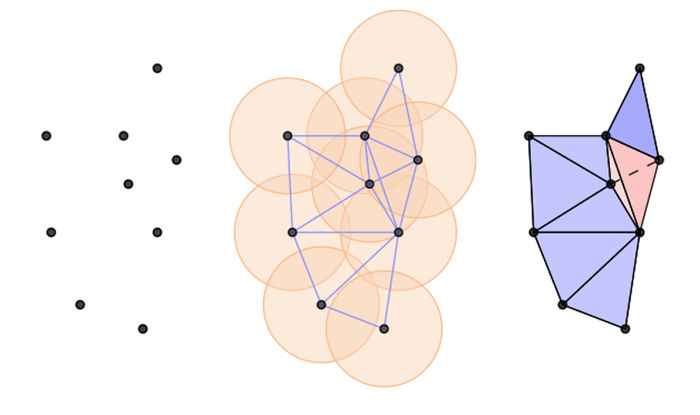
What’s next?
As part of the ELLIS honors program, Floor is continuing his research on topological deep learning under the supervision of Dr. Francesco Di Giovanni (Cambridge), Prof. Michael Bronstein (Oxford), and Dr. Erik Bekkers (UvA). His current interests are in extending the existing ideas in topological deep learning to a generative setting, e.g. for drug discovery.